Empowering Viral Genomic Surveillance in West Africa
In a leading-edge effort, the Celesta project enhances public health response in Guinea and Nigeria through advanced metagenomic sequencing setup.

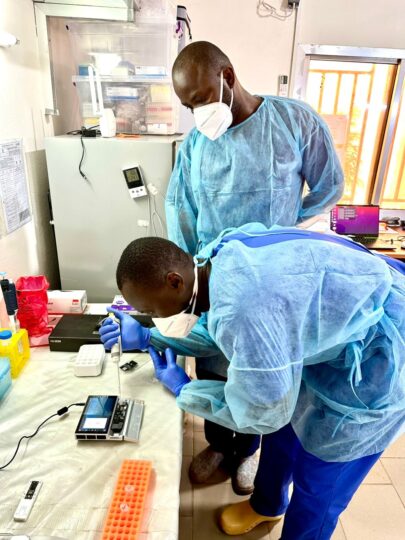
The ongoing threats posed by highly infectious viral pathogens necessitate versatile approaches to surveillance strategies. This particularly concerns regions of endemicity or at risk of virus emergence. Nanopore next generation sequencing offers such opportunity to setup molecular epidemiology capacities where needed. In 2024, three partner laboratories in Guinea1,2 and Nigeria3, comprising ten laboratory staff, benefited from a 1-year intensive nanopore metagenomics sequencing training. Furthermore, there are now two labs emerging as key genomic surveillance laboratories in their respective countries. One lab in Guinea is still undergoing a final setup. The full training program included both wet lab and bioinformatics. It equipped participants and labs with essential skills, equipment and reagents for nanopore sequencing. As result, teams are now empowered to conduct sequencing operations self-sufficiently. This includes basic on-site bioinformatics, offering local expertise to public health surveillance systems.
This newly acquired capability at the Centre de Recherche en Virologie, Laboratoire des Fièvres Hémorragiques Virales de Guinée (CRV-LFHVG) in Conakry has led to notable advances in disease surveillance. This especially concerns the timely detection and molecular identification (historical Clade IIa) of a human mpox case making it the first ever reported in Guinea to date (see article). It also supported both the timely diagnosis and further sequencing of several dengue, Lassa and yellow fever sporadic cases. In Nigeria it proved invaluable for understanding Lassa virus transmission dynamics during the epidemic peak in early 2024 (see article). Remote and on-site mentoring remains a key aspect of Celesta and ongoing work aiming at improving nanopore bioinformatics tools are ongoing.
The Celesta project marks a significant step forward in enhancing regional health security to infectious disease outbreaks in low- and middle-income countries. In so doing, it ensures that countries are better equipped to address future challenges. The innovation arising from this program also allowed for mobile nanopore sequencing development, and it is now integrated within the European Mobile Laboratory (EMLab) as a rapid response mobile laboratory to support in outbreak response. The efforts underscore the importance of sustainable scientific collaboration and local capacity development in the fight against infectious diseases. GHPP funding has been key in supporting our long-term collaboration leading to this year achievements.
1 Centre de Recherche en Virologie, Laboratoire des Fièvres Hémorragiques Virales de Guinée (CRV-LFHVG), Conakry, Guinea
2 Laboratoire des Fièvres Hémorragiques Virales de Gueckédou (LFHV-GKD), Gueckédou, Guinea
3 Irrua Specialist Teaching Hospital (ISTH), Irrua, Nigeria