SARS-CoV-2 Sequencing in Mozambique – First Local Training in “Genotyping of SARS-CoV-2”
Next-generation sequencing (NGS) analyses of SARS-CoV-2 genome is a crucial tool for monitoring of the pandemic worldwide. However, Mozambique lacks the local capacity to perform viral NGS. To contribute to change this reality, this project aims to establish workflows for SARS-CoV-2 whole-genome NGS locally.

In the first half of November 2021 four members of the technology implementation team of Research Center Borstel – Leibniz Lung Center (RCB) visited the National Institute of Health (INS) in Maputo, Mozambique to hold a joint training in “Next Generation Sequencing for prediction of drug resistance in clinical M. tuberculosis strains and genotyping of SARS-CoV-2”. Herein is reported the part of the workshop that focused on the sequencing of SARS-CoV-2 and subsequent bioinformatic analyses.
Since the beginning of the pandemic, dated from December 2019, knowledge generated by NGS data from circulating virus variants has contributed to drive containment measures worldwide. However, many resource-limited countries lack in local capacity for performing NGS of viral genomes.
The first PCR-confirmed COVID-19 case in Mozambique was announced on 22 March 2020, since then the country had relied on shipment of samples to other countries to perform SARS-CoV-2 sequencing. With this, in Mozambique it was usual to wait for weeks, or even months, before having data on the circulating viral variants. Thus, limiting the country’s and world response to the pandemic.
Supported by funds of the GHPP, the COVIDSeq project realized the first local training on NGS genotyping of SARS-CoV-2 in November 2021. PhD students and technicians of the INS were trained during one week in the state-of-the-art protocols used at RCB for SARS-CoV-2 surveillance in Germany. A Train the Trainer component was also included.
On-site trainings have a major role in the implementation of new technology since these activities allow not only to train local personnel, but also to adapt the workflows and to validate our protocols using the local infrastructure. The visit was also important to strengthen the cooperation between INS and RCB. Meetings with scientists, lab manager and stakeholders to exchange technical aspects as well as scientific discussions of data received were therefore also part of the agenda.

The training was based on a comprehensive theoretical explanation associated with a practical hands-on activity on every step of the workflow for SARS-CoV-2 whole-genome NGS. A summary of the training program can be seen below:
- Practical part: Selection of SARS-CoV-2 samples
- Lecture: Using NGS for SARS-CoV-2 analysis
- Practical part: RT of SARS-CoV-2
- Practical part: RT and PCR of SARS-CoV-2
- Practical part: Library prep
- Practical part: genome sequencing of SARS-CoV-2 using the iSeq100 platform
- iSeq run 2: SARS-CoV-2 samples
- Lecture: SARS-CoV-2 data analysis an interpretation
- Practical part: SARS-CoV-2 data analysis an interpretation
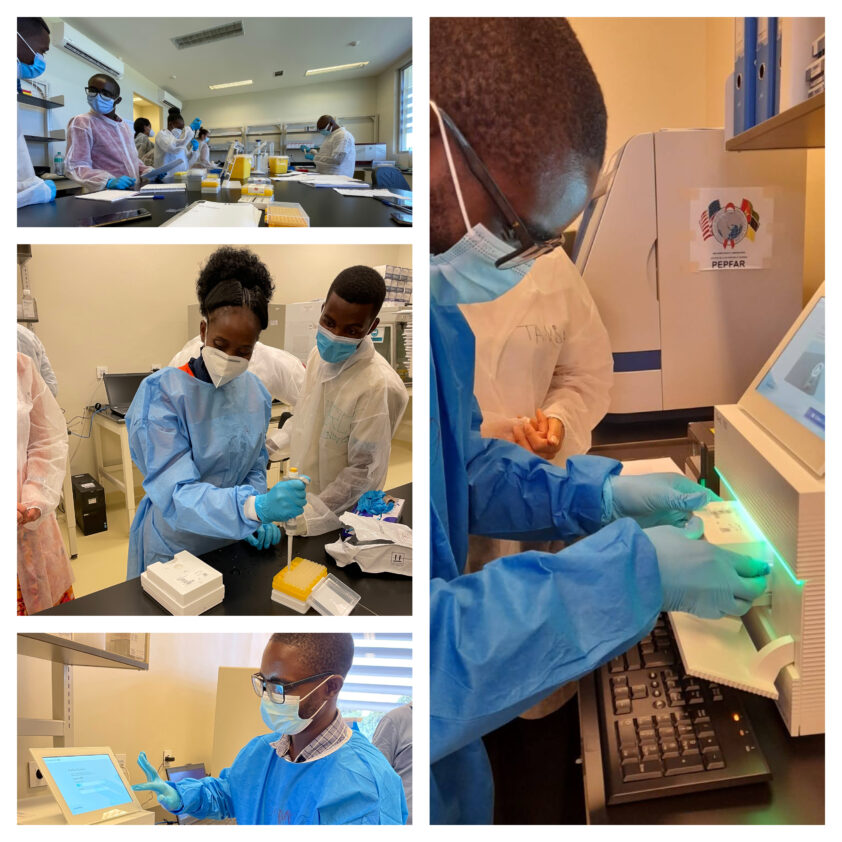
Trainees processed specimens collect from COVID-19 patients from Mozambique as well as standard samples brought from the Research Center Borstel to validate the workflow. The first iSeq run of SARS-CoV-2 samples done using local infrastructure and personnel was successfully finished on 11 November 2021. Data analysis and troubleshooting were also part of the training (as seen in the program).
As a next step, the trainees will perform the same protocols on their own (devices and consumables needed are provided by funds of the GHPP), and continuous support will be provided by phone, e-mail, and instant messaging apps. And a second site visit is planned for the beginning of 2022.
Ongoing whole-genome epidemiology analyses of SARS-CoV-2 in multiple other countries is being used for the monitoring of various other aspects of the pandemic worldwide, which includes diagnostics, genetic diversity/viral mutations, epidemiological pattern, and rational designs of therapeutic targets and vaccine. As a matter of fact, a recent discovery of a new Variant of Concern (VoC) Omicron circulating in South Africa was reported to the World Health Organization on 24 November 2021. South Africa shares borders with Mozambique, thus local sequencing capacity can help to elucidate if this new VoC is circulating also inside their country.
Despite the challenges faced with delivery of lab material and travel restrictions, Mozambique is now part of the group of countries with capacity for SARS-CoV-2 sequencing and their data will contribute to monitoring the pandemic outcome in that part of the globe.
Date: December 2021



