Modern Diagnostic of Drug-Resistance in TB – Training in “Next Generation Sequencing for Prediction of Drug Resistance in Clinical M. Tuberculosis Strains”
Tuberculosis (TB) is a widespread infectious disease associated with low social-economic-sanitary conditions. Upon treatment TB is usually curable. However, inaccurate diagnostic with the prescription of the wrong therapies may lead to relapse and resistance. Sequencing technology may improve TB diagnostic by allowing the prescription of patient-tailored treatments.

In the framework of the GHPP project SeqMDRTB_NET the technology implementation team of Research Center Borstel – Leibniz Lung Center (RCB) visited the National Institute of Health (INS) in Maputo, Mozambique in November 2021. INS has potential to set up the new technology of targeted next generation sequencing (tgNGS), which allows the elucidation of so called bacterial resistomes. A resistome specifically indicates all known antibiotic resistances of the TB causative pathogen Mycobacterium tuberculosis complex (Mtbc) present in a certain sample/patient.
With the help of tgNGS technology particular regions of the genome of Mtbc, that are known to be associated with the development of resistance, can be traced and the knowledge of this resistance conferring mutations can be used to outline a tailored therapy with higher probability of cure for the certain patient. During the visit, the technology implementation team also performed an optimization of the technology to local conditions, further adaptation of standard operational protocols (SOP), meetings with stakeholders, scientific discussions of preliminary data, the establishment of the NGS capacity, and a local training on tgNGS-technology.
Site visits and on-site trainings are crucial steps to understand our collaborators’ reality and to validate the use of new technologies in individual settings. Thus, allowing to form a competence network locally and between the partner institutions.

The first training in “Next Generation Sequencing for prediction of drug resistance in clinical M. tuberculosis strains and genotyping of SARS-CoV-2” comprised all essential steps of the technique, data analysis, and basic troubleshooting, also a Train the Trainer component. The current worldwide pandemic was also taken into account, as part of the workshop was dedicated to training on whole-genome genotyping of SARS-CoV-2, which will be described separately in another action report.
In total six participants, including local PhD students and technicians, attended the training (see program below)
- Welcome Addressed by Prof. Sofia Viegas (INS), Prof. Stefan Niemann (RCB)
- Course Introduction/Program adjustments/Meeting planning
- Laboratory Set Up
- Practical part: Selection of samples and DNA precipitation from GenoLyse
- Theoretical part: Documentation, discussion of SOPs and Protocols for the next days
- Practical part: Deeplex® Myc-TB PCR (DNA Dilution and multiplex PCR)
- Practical part: Clean up and preparation of Deeplex amplicons for NGS library generation
- Practical part: Deeplex NGS library preparation
- Practical part: NGS library quality control and starting the iSeq sequencer
- iSeq run 1: Deeplex samples
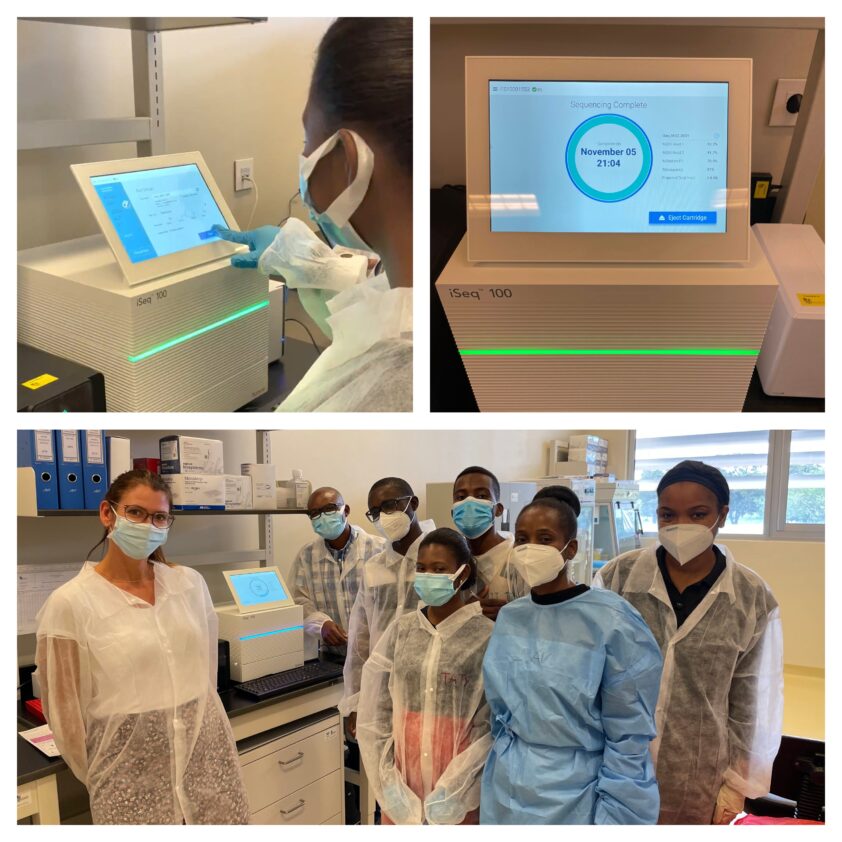
The trainees processed samples collected from Mozambican patients with confirmed drug-resistant TB and control samples brought by the RCB team to evaluate the workflow adapted to the local conditions. The first iSeq sequencing of TB samples was successfully performed on 5 November 2021.
All essential workflows and SOPs are now in place for the project and we expect the first patient samples to be included in the pilot study soon. As the next step, there are consumables available so that the training of participants can carry out the established protocols by themselves (devices and consumables needed were already provided by funds of the GHPP), but with ongoing support via cell phone, e-mail, and instant messaging apps.
As drug-resistant tuberculosis is an increasing threat to the health of the population in Mozambique, we hope that this new technology of tgNGS will help to increase the cure rate of patients with drug-resistance TB in the future. Furthermore, this GHPP project can be an important tool to train local staff and thus make the introduction of modern diagnostic tools in Mozambique sustainable.
Date: December 2021


